Work by Joel Lapin deals with structure fitting of orientationally dependent dipolar couplings and chemical shifts and automated algorithms for NMR spectroscopic assignment

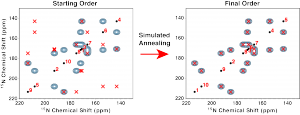

Panels a) and b) above show automated reassignment of experimental Pf1 phage solid-state NMR data.
Recently, we have been working on de novo design of NMR pulse sequences from first principles using GPU computing and simulated annealing. This method is termed ROULETTE which stands for “Random Optimization Using Liouville Equation Tailored To Experiment”. This optimization method allows one to considerably extend the FID signal, which dramatically narrows down the NMR spectral lines.
