Development of efficient pulse sequences is especially important for solid-state NMR, where the dipole-dipole interactions among the spins are not averaged out (unlike in solution NMR), which would result in significant line broadening. Here we focus on developing high-resolution pulse sequences correlating 1H, 13C, and 15N spins for the purpose of spectroscopic assignment and measurements of the orientationally dependent HC and HN dipolar couplings for structure determination of membrane proteins in aligned planar lipid bilayers.


Work by Rob Knox and Wenxing Tang established purely spectroscopic assignment in NMR of aligned samples using the 15N-15N and 1H-15N correlation experiments shown above.

Work by Sophie Koroloff illustrates the use of selective excitation pulses for initiating spectroscopic assignment of crowded regions in the solid-state NMR spectra of Pf1 coat protein reconstituted in magnetically aligned bicelles.


Recent work by Emmanuel Awosanya makes use of T1rho relaxation anisotropy in aligned perpendicular bicelles to extract rotational diffusion coefficients of membrane proteins in liquid-like lipid bilayers. A. Spectrum of Pf1 coat protein with color-coding for the individual peaks. B. Individual T1rho decay curves for the select peaks. C. Linear regression plots allow one to extract the diffusion coefficient from fitting the data to a relaxation model (cf. Awosanya et al., Biophys. J. 114, 392–399, 2018 for further details).
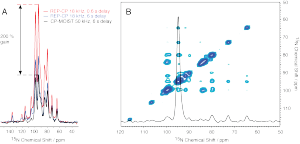
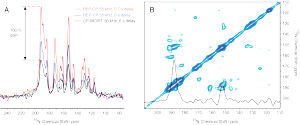
Work by Deanna Tesch and Sophie Koroloff employs paramagnetic relaxation enhancement and repetitive (multiple-contact) cross polarization to dramatically enhance NMR sensitivity. Shown above are spectra of Pf1 reconstituted in unflipped (perpendicular) and flipped (parallel) bicelles with the corresponding sensitivity gains obtainable by the method.